K. Ashok Kumar*, P. Somasundaram,
G.K. Rajesh, R. Chandrasekar, N. Balachandran and V. Sivaprasad
The 1st, 2nd,
4th, 5th and 6th authors are with Central Silk
Board (CSGRC, Hosur) India. The 3rd author is with Council for
Nature Conservation and Environmental Protection (CONCEPT), India
Introduction
The silkworm Bombyx mori L. is one of the earliest examples of insect
domestication and commercial exploitation. The insect is believed to be first
identified and put to use by the Chinese nearly 4500 years ago. Since then the
species has been subject to severe selective pressure, targeting improved
quantitative traits (with reference to silk). As a result the cocoon shell weight
has increased manifold while the insect has lost many of its defensive
features that once helped it survive in the wild fighting whether, natural
enemies and diseases. The fact that one of its closest ancestors namely Bombyx mandarina still lives in the wild
enables us comparison which testifies to the above statements. While the
progress so far made is quite impressive (in human angle), it is widely perceived
that the insect leaves much to be desired in terms of quantitative output and
tolerance to adverse environment, pests and diseases. Thus location specific
hybridization programs are in vogue towards evolving productive hybrids which
are not only tolerant to specific micro climates and pathogenic flora but also
adaptable to resource saving techniques such as artificial diet. The process has lead to creation of silkworm
germplasms in many countries offering a wide variety of genetic resources to
select from. However, selection of parental breeds has become quite a
challenging task, given such a vast and varied collection of geographic races,
evolved breeds, transgenic lines, mutant stocks etc. This article sums up a
series of studies conducted at the Central Germplasm Resources Centre (CSGRC)
Hosur, India and proposes the application of protein markers for
characterization of silkworm genetic resources to identify duplicate genotypes
and to find out intra and inter racial variations.
Experimental material
This
paper summarises a few laboratory screening studies conducted on 28 silkworm
accessions (14 multivoltine races and 14 bivoltine races) maintained at Central
Sericultural Germplasm Resource Centre (CSGRC), Hosur Tamil Nadu, India (latitude
12°45’N and longitude 77°5’E,
altitude 942M AMSL). The center maintains a total of 421 accessions: 71
multivoltines and 350 bivoltines (Thangavelu et al., 1997 and 2000). The
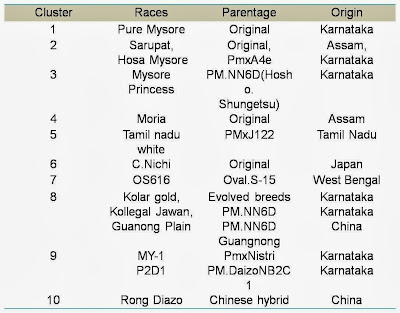
details of the races used for study; such as geographical origin, and cluster
groups based on genetic distances are furnished in tables
1-3
 |
Table 1. Details of geographical origin and
class of 28 silkworm accessions
|
 |
Table
2. Cluster groups based on genetic distances in 14 MV races
|
 |
Table
3. Cluster groups based on genetic distances in 14 BV races
|
Biochemical markers for characterization
Biochemical markers for hardiness
Amylase
enzyme as a biochemical marker was studied in silkworm genetic resources for
its effective usage as surrogate breeding parameter to correlate amylase
activity with economic traits. This study was aimed at identification of
suitable biochemical markers for the successful implementation of Marker
Assisted Selection (MAS) which being used in other fields of breeding (Datta,
1998, Jayaswal et al., 2000). Out of a number of biochemical parameters viz.,
digestive amylase, invertase, protease, alkaline phosphatase and haemolymph
trehalose, only the digestive amylase has a significant positive correlation
with survival on one hand and negative correlation with weight of larvae,
cocoon and shell on the other (Chatterjee et al., 1992). The genetic divergence
in term of activity as well as isozyme, polymorphism coupled with its role in
better digestibility and survivability indicates the prospect of using amylase
as a marker in silkworm breeding. It is reported that multivoltine races
contain dominant amylase gene (Ae+) with four to five band polymorphism
responsible for inducing hardiness in multitovltine races. This gene (Ae+)
recessive in temperate races with no amylase isozyme band (Null type) may be
responsible for less tolerance to climatic fluctuations. Thus the dominant (Ae+) gene can be
transferred to temperate races through a suitable vector for developing hardy
bivoltine races. Similarly haemolymph amylase isozyme studies conducted so far
showed variability in its activity in different silkworm stocks. Most of the
hibernating high yielding races of Japanese and European races are homozygous
for Amy dn Detailed characterizations of silkworm germplasm stocks
are vital for the effective use in potential breeding resources. Hierarchical
agglomerative clustering by UPGMA method using Euclidean distance of 54 stocks
on the basis of yield attributes alone resulted in 7 clusters indicating the
significance of such data in the characterization and classification of
silkworm germplasm stocks(Chatterjee and Datta, 1992). The categorization of
silkworm strains into five yielding groups on the basis of discrimination
function scores of economic characters and esterase isozyme activity at
different developmental stages was done on twenty silkworm genotypes (Ram and
Lal, 2002).
Biochemical marker for genetic variations
Studies
carried out on biochemical characterization of silkworm races of different
origins from germplasm stocks with four metabolic enzymes viz., a
esterase, b esterase , acid phosphatase, and
alkaline phosphatase were able to compare and relate the genetic relationship
among 14 MV races viz., Pure Mysore,
Sarupat, Moria, Tamil Nadu white, C.Nichi, Hosa Mysore, Mysore Princes, Kolar
Gold, Kollegal Jawan, MY-1, P2D1, Rong Daizo, Guangnang and OS-616 and 14 BV races viz., Alps jaunne, Alps
yellow, Cevenesse yellow, Ascoli yellow, Meigtsu, B-36, B-37, B-39, B-40,
J-112, J-122, Yakwei, Changnaung and C-122. The results on electrophoretic
variation on isozyme profiles are seen in the band mobility (Fig. 1a,b).
 |
Fig.1a.
Genetic polymorphism on esterase isozyme among 14 MV races. Various
bands designated as A,B,C & D.
|
 |
Fig.1b.
Genetic polymorphism on esterase isozyme
among 14 BV races. Various bands designated as A,B,C & D.
|
A
total of 4 bands was observed in multivoltine races and 7 bands in bivoltine
races. All the observed 4 bands in multivoltine and 7 bands in bivoltine were
not present uniformly in all 28 races studied. These variable bands present in
them were identified as race specific markers and analysed through pop-gene
software package for genetic analysis (Yeh et al., 1999). Thus 9 clustering groups were identified
among 14 multivoltine races and 8 clustering groups among 14 bivoltine
races.
The clustering helps us to relate their origin,
racial characters and the parentage of the evolved breeds. Among 14 MV silkworm
races studied, higher genetic distance was seen between C.Nichi (Original race
of Japan origin) and Rong Daizo (Evolved breed of China origin). Similarly in
BV silkworm races, such higher genetic distance was observed between B-36
(Japan origin) and C-122 (China origin).
A detailed study on biochemical characterization in these line and
proper classification of germplasm stocks is vital for the effective use in
potential breeding resources. The genetic diversity and population genetic
structure of twelve silkworm races revealed in the present study would be
utilized for an effective conservation plan and breeding strategies. Based on
the results observed in the present study, it is inferred that populations of
silkworm races J-112 and NB4D2 would be very useful in a breeding programme,
because their higher genetic diversity and alleles.
Storage protein profiles for
genetic variations.
Studies on storage
protein expression in the final instar larvae of silkworm races help to
identify and cluster them based on the genetic variability in storage protein
profiles. Storage protein variation was studied in eleven popular multivoltine
silkworm (Bombyx mori) breeds encompassing different parentage and
origin, as furnished in table 4. The main
feature of storage protein variation in the multivoltines is found to be inter-origin
variability in unit area expression of storage protein. The storage protein
levels (SP-2) among these popular breeds as seen in SDS-PAGE also differed as
evident from the SDS PAGE profiles in Fig. 2.
 |
Table 4:Variability in protein expression of storage
protein (SP-2) level in
multivoltine Silkworm breeds of Bombyx mori |
 |
| Fig.2. Genetic polymorphism on storage proteins in 11 MV silkworm races |
Based on densitometry scanning of SDS-PAGE bands of storage protein levels, six clustering of eleven races was done using Ward’s Minimum Variance clustering analysis. Storage protein has an important role to play on metamorphic features of larval weight and pupation rate of B.mori as evident from correlation factors. Thus study on genetic variability existing among storage protein level in silkworm breeds may be useful to cluster the silkworm races into different clusters having similarity in the storage proteins and their effect on growth traits.
Heat stable esterase isozyme profiles for thermo tolerance
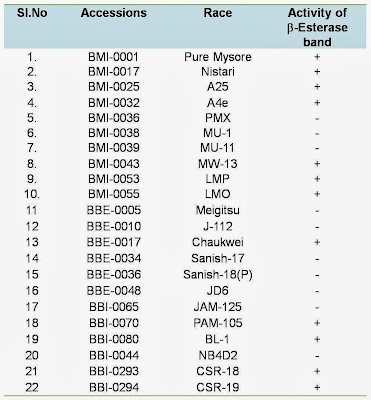
The climatic condition of tropics, particularly in summer is not conducive to rear high yielding bivoltine hybrids. Keeping this in view, characterization of silkworm breeds for thermo tolerance is imperative in order to utilize such those identified thermo tolerant breeds in the breeding plan to evolve productive bivoltine /multivoltine breeds/hybrids. Genetic dissection of silkworm races carried out by several Japanese scientists (Sarkar, 1998) has indicated that certain enzymes are responsible for hardiness and related quantitative characters. Wu et al (1993) reported that the thermo tolerance varies with strains and is positively relative to the activity of a heat stable esterase (HsEST) analysed in larval body or midgut. This enzyme activity can be used as an indicator for the silkworm thermo tolerance. Further, there is always a heritable linkage between characteristics of high temperature tolerance and disease resistance (Samson and Chandrashekaraiah, 1998). In view of this, breeds with such trait should be characterized in the germplasm as resource materials for developing thermo tolerant breeds/hybrids. Non specific b-esterase band (Est 3) in haemolymph of CB5 (GP) and its syngenic lines among the silkworm stocks withstood a temperature up to 80 ±° C for 10 min indicating naturally available thermo stable esterase protein in the haemolymph of thermo tolerant races (Chattopadhyay et al., 2001). With this main objective, the presence of heat stable esterase isozyme band activity was studied in selected silkworm races along with thermo tolerant races CSR-18 and CSR-19 to identify such those breeds which can withstand higher temperature prevailing in some of the areas where during summer the temperature may shoot up to 40 °C and above. A total 10 MV races viz. Pure Mysore, Nistari, A25, A4e, PMX, MU-1, MU-11,MW-13, LMP and LMO and 12 BV races viz., Meigitsu, J-112, Chukwei, Sanish-17, Sanish-18 (P), JD6, JAM-125, PAM-105, BL-1, NB4D2, CSR-18 and CSR-19 has been studied for heat stable esterase isozyme activity to relate thermal tolerance in the above silkworm races. Heat stable esterase isozyme study showed the presence of active b-esterase band in multivoltine silkworm races of Pure Mysore, Nistari, A25, A4e, MW-13,LMP and LMO and in Chaukwei, PAM-105,BL-1,CSR-18 and CSR-19 of bivoltine silkworm races. Such active b-esterase band was absent in multivoltine races of MU-1, MU-11 and PMX and in Meigtsu, J-112, Sanish-17, Sanish (18), JD-6, JAM-125, NB4D2 in bivoltine silkworm races. The presence of active b-esterase band in the haemolymph of silkworm races may be responsible for thermo tolerance. The differential Esterase isozyme band activity observed in 11 multivoltine and bvoltine breeds is furnished in table 5.
 |





No comments:
Post a Comment